Last month a new R package was released called ODBC to improve the ability to connect to relational databases. By using these universal connection packages such as DBI and ODBC, you’ll be able to easily port your code to different databases. If you currently use RODBC to connect to Oracle, you’ll like ODBC more! ODBC is a completely rewritten package from RODBC. The creators state that it’s been well tested on SQL Server, PostgreSQL and MySQL. I found some issues with using it against an Oracle source but not enough to keep me from using it.
The benefit of using this ODBC connection is that you can now push R commands and processing to the database instead of running in your R instance. When working with large datasets, this will speed up the amount of time your R code takes to run by using database server resources instead of your own machine or R server.
Connecting to the Oracle Database
To start, you’ll need an Oracle ODBC connection setup on your computer. That requires an Oracle client on your machine to be able to set it up. I’m not going to walk through setting this up but you can find some help here.
The ODBC package uses the DBI package to connect to the database. DBI is a common interface between R and DBMS systems and has also recently been updated. You’ll need to install both the DBI and the ODBC packages and load them into your environment.
install.packages("DBI")
install.packages("ODBC")
library(DBI)
library(odbc)Next we’ll setup a connection in R to the ODBC data source. My ODBC data source name is oracledb as you’ll see below. Note if you are not using RStudio, you’ll need to find another secure method for storing your password. If you are using RStudio, install the rstudioapi package to be able to use a prompt for the database password when connecting. It’s also possible to use the keyring or config packages to pull from your operating system’s stored passwords.
con <- dbConnect(odbc::odbc(), "oracledb", UID="samples", PWD= rstudioapi::askForPassword("Samples User Password"))The connection to the Oracle database is giving me an error about a feature not being implemented but I’m still able to query it successfully so I just ignore this message.

Once connected, you can use DBI methods to read tables or show the tables available:
#Show all tables this user can view in this schema dbListTables(con, schema = "SAMPLES")

If you get a response of character(0) when you try to list the tables, check the case of the schema you entered. If I use lower case for SAMPLES in the code above, I’ll get no tables returned and character(0) instead.
DPLYR Improvements with ODBC
When you use an ODBC source with DPLYR & DBPLYR methods, R will translate your R code to SQL and run against the database without pulling the data into your R environment. This is awesome because when you are working in R it is typically with large datasets that are difficult to use on your local machine or R instance.
Using that connection that we created earlier called “con”, we’ll first create a reference to our ORDERS table in the Oracle database.
db_orders <- tbl(con, "ORDERS")
We can view the first six rows of this table with head and then just to prove it’s running in the database, we can view the SQL created with the show_query method. You’ll notice that the SQL created is specific Oracle syntax. If you run this against postgres or SQL Server or others, it’ll be specific to those databases.
head(db_orders) show_query(head(db_orders))
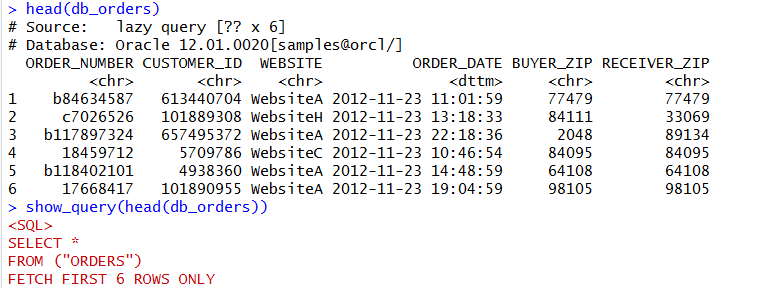
We can use the DBPLYR package to put together SQL syntax for querying. While the commands are not identical to SQL, they are fairly logical and easily translate.
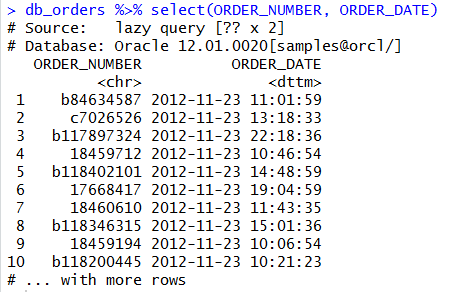
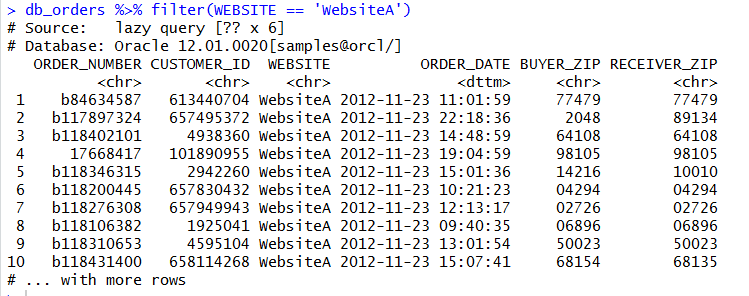
We can aggregate and use grouping and ordering also:
show_query(db_orders %>% group_by(WEBSITE) %>% summarize(order_count = count()) %>% arrange(WEBSITE))
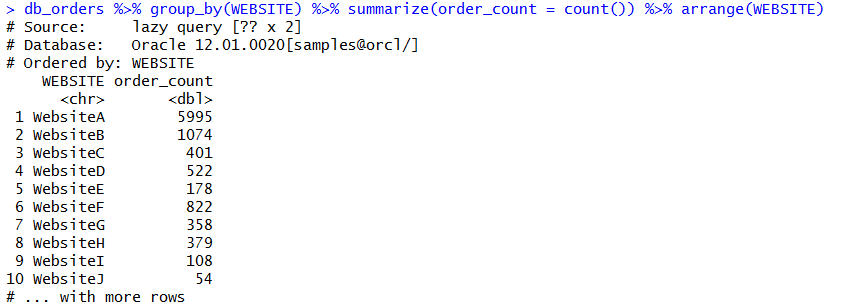
You can check the SQL of this statement to evaluate the quality of the SQL that R is building. There’s two ways we can do this. We can paste our R DBPLYR code into a show_query method. Or, we can save our pre-built code into a variable that we can reference with show_query.
show_query(db_orders %>% group_by(WEBSITE) %>% summarize(order_count = count()) %>% arrange(WEBSITE))
order_count_by_site <- db_orders %>% group_by(WEBSITE) %>% summarize(order_count = count()) %>% arrange(WEBSITE)
order_count_by_site %>% show_query()

It’s possible to use the SQL that R is creating to extract a dataset from the database and store in your R environment using the collect method. Keep in mind that this could take a while if the dataset is large.
order_count_site_download <- order_count_by_site %>% collect()

The data transfer can also work the other way and upload data into the Oracle database using the copy_to method. Below I’m referencing my R data frame called order_count_site_download and asking it to create a new table in the database called ORDER_COUNT_AGG with the data in the data frame.
copy_to(con, order_count_site_download, "ORDER_COUNT_AGG",
temporary = FALSE,
indexes = list(WEBSITE)
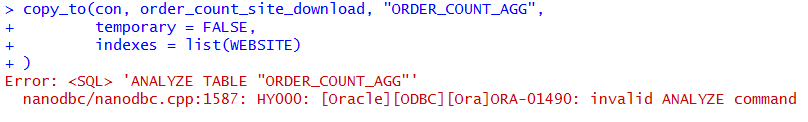
)The copy_to method is also able to create indexes on the table. However, I found that the Oracle implementation of this table creation throws an error when the method tries to analyze the table. In my environment the table was created with the data in it but the table does not have statistics gathered or indexes.
copy_to(con, order_count_site_download, "ORDER_COUNT_AGG",
temporary = FALSE,
indexes = list(WEBSITE)
)
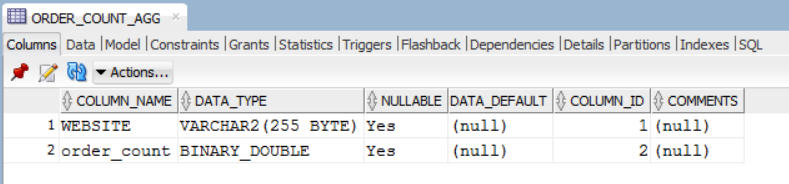
We can view this new table in R using the DBI method dbReadTable.
dbReadTable(con, "ORDER_COUNT_AGG")
There are more packages and methods which can leverage this ODBC connection so that you can run R code in the database. This is only the beginning!
